Quickstart
This chapter aims to explain how to use GalaxyChop in general. Here you can see a simple example of
Reading a galaxy from a file HDF5.
Explore the attributes of the galaxy.
Some graphics.
Aligning and centering the galaxies.
A simple decomposition.
1. Conceptual overview
By a design decision, and after several prototypes, GalaxyChop was designed in two layers
A data representation level, where we defined a class
Galaxywhich encapsulates all the physical properties useful for the dynamic decomposition.A
modelslevel, which understands theGalaxyinstances and returns an instance of typeComponentswhich represents “to which part of the galaxy each stellar particle corresponds to”.
This first tutorial aims only to give an overview of the project and not to go into too specific details.
Note: About the content
This tutorial is not intended to be a scientific explanation of why and how to perform a dynamic decomposition of galaxies, only to explain the functionalities of GalaxyChop.
2. Creating your first Galaxy
GalaxyChop represents galaxies with a class called galaxychop.Galaxy, which is a collection of gas, dark matter and stars particles each characterized by 3 positions, three velocities, masses and various energies.
To create a Galaxy type object, there are several alternatives, the most useful being to read all the data from a file type HDF5.
So first of all it is necessary to obtain a file with the appropriate content. It is recommended to download this file gal394242.h5 and place it where it is convenient for you.
Note
For convenience we have placed the file gal394242.h5 in the same folder where this tutorial is located.
As a first step it is necessary to import the GalaxChop library, and by simple habit we have taken the practice of assigning to the library the alias gchop.
[1]:
import galaxychop as gchop
Next we load the file gal394242.h5 using the read_hdf5 function. This will generate an instance of the gchop.Galaxy class.
[2]:
gal = gchop.read_hdf5("gal394242.h5")
gal
[2]:
Galaxy(stars=32067, dark_matter=21156, gas=4061, potential=True)
3. Galaxy attributes
Galaxies internally consist of three sets of particles , one for stars, one for dark_matter and one for gas.
All these sets can be accessed, and expose a set of physical properties
For example, if we wanted to access the positions (x, y, and z) of the stellar particles:
[3]:
gal.stars.x, gal.stars.y, gal.stars.z
[3]:
(<Quantity [ 0.12017286, -0.01992682, -0.09766993, ..., 15.06957949,
17.21394372, 9.28556454] kpc>,
<Quantity [ 1.51520602e-03, 3.25905789e-02, -2.27875536e-02, ...,
4.52400820e+00, -1.96094867e-01, 1.15665035e+01] kpc>,
<Quantity [-3.73908514e-02, 5.12012087e-03, 4.66359649e-02, ...,
1.15501605e+01, -1.40433638e+01, 7.57282498e+00] kpc>)
It can be observed that the positions have units which can be bypassed using the arr_ accessor.
For example if we would like to access the gas masses (m) but without units:
[4]:
gal.gas.arr_.m
[4]:
array([2579202.70087197, 1368584.63520184, 1392260.60127839, ...,
990849.80320185, 1456959.43501778, 1760535.66974588])
The galaxy instances, on the other hand, have useful methods to summarize the data from the three sets, such as summarize the data coming from the three sets, such as angular_momentum_ or gal.kinetic_energy_.
Finally both the arrays and the galaxy in general have methods to create Pandas DataFrames with the particle data.
[5]:
gal.to_dataframe()
[5]:
| ptype | ptypev | m | x | y | z | vx | vy | vz | softening | potential | kinetic_energy | total_energy | Jx | Jy | Jz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | stars | 0 | 5.224283e+05 | 0.120173 | 0.001515 | -0.037391 | 10.773575 | -6.878906 | -20.425400 | 0.0 | -195699.620206 | 290.293111 | -195409.327095 | -0.288157 | 2.051746 | -0.842982 |
| 1 | stars | 0 | 9.745897e+05 | -0.019927 | 0.032591 | 0.005120 | 20.282349 | 8.661957 | -7.947495 | 0.0 | -196176.962277 | 274.782915 | -195902.179362 | -0.303364 | -0.054520 | -0.833619 |
| 2 | stars | 0 | 6.935776e+05 | -0.097670 | -0.022788 | 0.046636 | -14.897980 | 6.957092 | -10.818886 | 0.0 | -195152.120168 | 193.699612 | -194958.420557 | -0.077915 | -1.751461 | -1.018987 |
| 3 | stars | 0 | 1.070959e+06 | -0.007224 | -0.138971 | 0.176169 | -8.665253 | -4.337433 | 5.506927 | 0.0 | -194695.767625 | 62.113089 | -194633.654536 | -0.001182 | -1.486768 | -1.172886 |
| 4 | stars | 0 | 6.013803e+05 | 0.095277 | 0.001167 | 0.100484 | 23.508469 | -7.842865 | -3.754723 | 0.0 | -195703.942688 | 314.128285 | -195389.814402 | 0.783702 | 2.719973 | -0.774688 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 57279 | gas | 2 | 1.400046e+06 | 2.415113 | -8.443658 | -0.427454 | 117.975723 | -243.209320 | 31.390686 | 0.0 | -129252.826165 | 37027.209909 | -92225.616256 | -369.012963 | -126.241204 | 408.768823 |
| 57280 | gas | 2 | 1.375867e+06 | -4.756847 | 5.970680 | -4.109694 | -136.718105 | 232.501099 | -56.024063 | 0.0 | -129835.893336 | 37943.648417 | -91892.244919 | 621.006707 | 295.371727 | -289.672123 |
| 57281 | gas | 2 | 9.908498e+05 | -9.376904 | -1.965296 | -0.563573 | -188.311616 | 31.467773 | 179.615128 | 0.0 | -132966.102621 | 34356.539847 | -98609.562774 | -335.262575 | 1790.361085 | -665.158420 |
| 57282 | gas | 2 | 1.456959e+06 | -6.880090 | 0.314529 | -6.284694 | -261.390251 | 69.243408 | -137.456593 | 0.0 | -132205.106762 | 46006.913910 | -86198.192853 | 391.939479 | 697.043885 | -394.186002 |
| 57283 | gas | 2 | 1.760536e+06 | -7.371887 | 2.670409 | -6.167000 | -282.899384 | 138.414307 | -134.297680 | 0.0 | -127609.955549 | 58613.224160 | -68996.731389 | 494.971200 | 754.613090 | -264.917457 |
57284 rows × 16 columns
4. Visualizing the galaxy
Galaxies have a set of utilities to visualize on the one hand all the particles in the real space; and on the other hand only the stellar ones in the circularity space.
Thus a good summary plot of a galaxy can be made with the command:
[6]:
gal.plot()
[6]:
<seaborn.axisgrid.PairGrid at 0x7f288e98baf0>
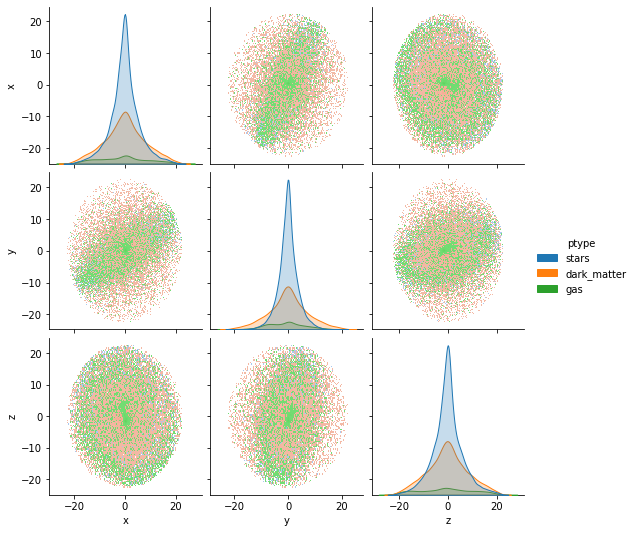
At this point we observe in the graph that the galaxy is neither centered nor aligned, this can be validated with:
[7]:
print("Centered:", gchop.is_centered(gal))
print("Aligned:", gchop.is_star_aligned(gal))
Centered: False
Aligned: False
To correct this GalaxyChop has the center() and star_align() functions which can be composed as follows
[8]:
gal = gchop.star_align(gchop.center(gal))
print("Centered:", gchop.is_centered(gal))
print("Aligned:", gchop.is_star_aligned(gal))
Centered: True
Aligned: True
We can also visualize the result, for example by comparing the positions at x and y, highlighting according to the particle type.
[9]:
gal.plot.hist("x", "y", labels="ptype")
[9]:
<AxesSubplot:xlabel='x', ylabel='y'>
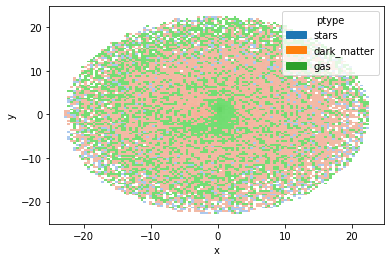
5. Chop the galaxy
Finally, let’s decompose a galaxy into its parts. The package offers several decomposition models as well as a framework to create new methods. methods.
For didactic reasons we will stick to the method presented in the work of Abadi et al.(2003), which in the context of GalaxyChop is called JHistogram.
The analysis can be summarized in the following steps
Instantiate the decomposer
[10]:
decomp = gchop.models.JThreshold()
Decompose the galaxy into components
[11]:
components = decomp.decompose(gal)
components
[11]:
Components(57284, labels=[ 0. 1. nan], probabilities=False)
As you can see by the labels [0 1 nan] the decomposer found two components (0 and 1) and could not classify at least one particle, so the meaning of nan for Galaxychop can be understood as “I don’t know what to do with this particle”.
Another detail to take into account is that GalaxyChop decomposes galaxies, but since the dynamic decomposition for gas and dark matter is not defined, it assigns the label nan to all particles.
Finally, it is seen that this classifier does not provide probabilities of particle belonging to a label as it does particle to a label as does GaussianMixture.
Now we can see the division of the particles in the circularity space.
[12]:
gal.plot.circ_pairplot(labels=components, lmap={0: "Spheroid", 1: "Disk"})
[12]:
<seaborn.axisgrid.PairGrid at 0x7f288576a7c0>
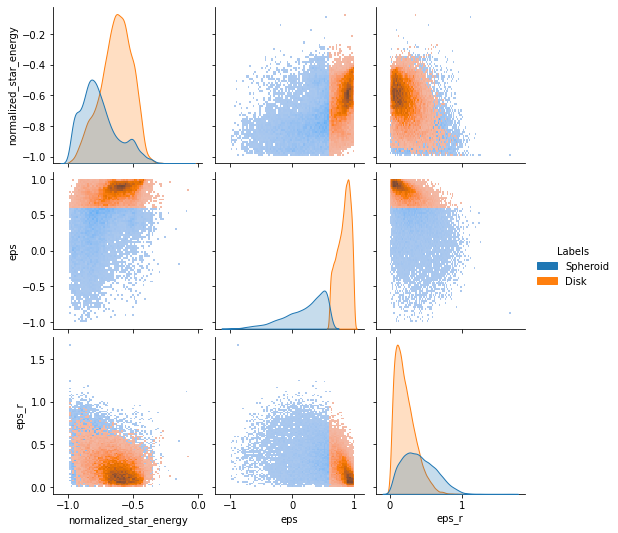
or in the real space
[13]:
gal.plot.pairplot(
labels=components,
lmap={0: "Spheroid", 1: "Disk"},
palette="inferno",
)
[13]:
<seaborn.axisgrid.PairGrid at 0x7f287e964fa0>
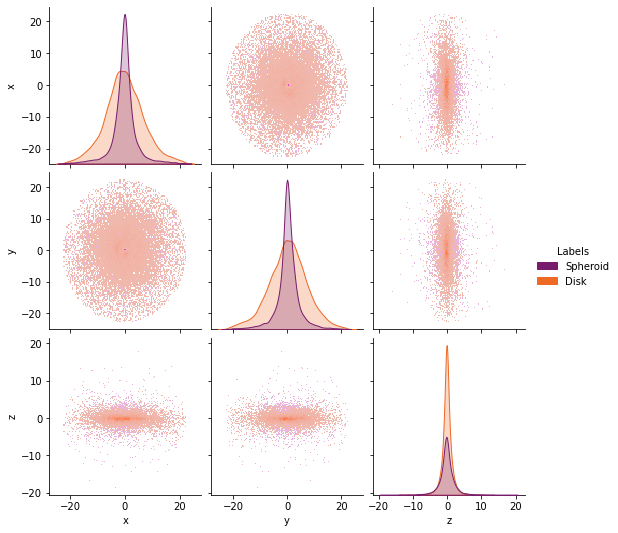
Finally you can convert the entire components into a Dataframe
[14]:
components.to_dataframe()
[14]:
| labels | ptypes | |
|---|---|---|
| 0 | 0.0 | stars |
| 1 | 0.0 | stars |
| 2 | 0.0 | stars |
| 3 | 0.0 | stars |
| 4 | 0.0 | stars |
| ... | ... | ... |
| 57279 | NaN | gas |
| 57280 | NaN | gas |
| 57281 | NaN | gas |
| 57282 | NaN | gas |
| 57283 | NaN | gas |
57284 rows × 2 columns